Advanced R - Notes (part2)
Advanced R (Hadley Wickham).
knitr::opts_chunk$set(error = TRUE)suppressPackageStartupMessages(library(tidyverse))
suppressPackageStartupMessages(library(rlang))
suppressPackageStartupMessages(library(lobstr))
suppressPackageStartupMessages(library(withr))
suppressPackageStartupMessages(library(glue))
# devtools::install_github("openpharma/simaerep@v0.3.1")
suppressPackageStartupMessages(library(simaerep))Metaprogramming
Basics
We are ignoring environments for now.
Capturing/Quoting an Expression or many expression
ex1 <- rlang::expr(mean(mtcars$disp))
ex1## mean(mtcars$disp)ex2 <- quote(mean(mtcars$disp))
ex2## mean(mtcars$disp)exs1 <- rlang::exprs(mean(mtcars$disp), median(mtcars$disp))
exs1## [[1]]
## mean(mtcars$disp)
##
## [[2]]
## median(mtcars$disp)exs2 <- alist(mean(mtcars$disp), median(mtcars$disp))
exs2## [[1]]
## mean(mtcars$disp)
##
## [[2]]
## median(mtcars$disp)Evaluating/Unquoting an Expression
eval(ex1)## [1] 230.7219lapply(exs1, eval)## [[1]]
## [1] 230.7219
##
## [[2]]
## [1] 196.3Capturing an Expression as a String - Deparsing
str_ex1 <- deparse(ex1)
str_ex1## [1] "mean(mtcars$disp)"rlang::expr_text(ex1)## [1] "mean(mtcars$disp)"Parsing Strings and Evaluating Code from Strings
parse(text = str_ex1)## expression(mean(mtcars$disp))eval(parse(text = str_ex1))## [1] 230.7219rlang::parse_expr(str_ex1)## mean(mtcars$disp)eval(rlang::parse_expr(str_ex1))## [1] 230.7219Converting Strings to Symbols
rlang::sym("foo")## fooas.name("foo")## fooSymbols to Strings
deparse(rlang::sym("foo"))## [1] "foo"rlang::expr_text(as.name("foo"))## [1] "foo"Code is a tree
Code can be displayed as an abstract synthax tree (AST) with functions/expressions/symbols as nodes and literals/constants as leaves.
lobstr::ast(f(a, "b"))## █─f
## ├─a
## └─"b"lobstr::ast(f1(f2(a, b), f3(1, f4(2))))## █─f1
## ├─█─f2
## │ ├─a
## │ └─b
## └─█─f3
## ├─1
## └─█─f4
## └─2lobstr::ast(1 + 2 * 3)## █─`+`
## ├─1
## └─█─`*`
## ├─2
## └─3Subsetting and modifiyng Expressions
length(ex1)## [1] 2ex1[[1]]## meanex1[[2]]## mtcars$dispex1[[1]] <- quote(median)
ex1[[1]]## medianex1## median(mtcars$disp)eval(ex1)## [1] 196.3Quoting/Unquoting in Functions
here we want to capture the user-supplied expression
capture1 <- function(x) {
quote(x)
}
capture1(mean(mtcars$disp))## xcapture2 <- function(x) {
rlang::enexpr(x)
}
capture2(mean(mtcars$disp))## mean(mtcars$disp)capture3 <- function(x) {
substitute(x)
}
capture3(mean(mtcars$disp))## mean(mtcars$disp)substitute() can also be used to alter expressions
substitute(mean(mtcars$disp), list(mean = quote(median), disp = quote(vs)))## median(mtcars$vs)bang bang !!
In rlang when constructing expression we can chose to selectively unquote parts of the expression using !! bang bang.
x <- rlang::expr(-1)
rlang::expr(f(!!x, y))## f(-1, y)a <- rlang::sym("foo")
b <- 1
rlang::expr(f(!!a, !!b))## f(foo, 1)bang bang bang !!!
selectively unquote lists of expressions using !!! bang bang bang.
xs <- rlang::exprs(1, a, -b)
rlang::expr(f(!!!xs, y))## f(1, a, -b, y)# Or with names
ys <- rlang::set_names(xs, c("a", "b", "c"))
rlang::expr(f(!!!ys, d = 4))## f(a = 1, b = a, c = -b, d = 4)Dynamic Dots …
rlang::list2() allows a function to use !!! and := with its ... argument, which unpacks the list.
f <- function(...) {
out <- rlang::list2(...)
rev(out)
}
x <- list(alpha = "first", omega = "last")
f(!!!x)## $omega
## [1] "last"
##
## $alpha
## [1] "first"f(x)## [[1]]
## [[1]]$alpha
## [1] "first"
##
## [[1]]$omega
## [1] "last"nm <- "foo"
f(!!nm := "bar")## $foo
## [1] "bar"Provide lists as function arguments
use rlang::exec() which uses dynamic dots
# Directly
exec(mean, x = 1:10, na.rm = TRUE, trim = 0.1)## [1] 5.5# Indirectly
args <- list(x = 1:10, na.rm = TRUE, trim = 0.1)
exec(mean, !!!args)## [1] 5.5# Mixed
params <- list(na.rm = TRUE, trim = 0.1)
exec(mean, x = 1:10, !!!params)## [1] 5.5- use
do.Call
do.call(mean, list(x = 1:10, na.rm = TRUE, trim = 0.1))## [1] 5.5Quosures
When evaluating an expression we can control the environment. Quosures consist of an expression and an environment.
They are similar to formulas in base R
Formulas
we can extract environment and formula expressions using rlang functions and evaluate both.
construct_formula <- function() {
env_x <- 3
~runif(env_x)
}
f <- construct_formula()
f## ~runif(env_x)
## <environment: 0x7fe60b693388>eval(rlang::f_rhs(f), envir = rlang::f_env(f))## [1] 0.4454028 0.3837579 0.3728023rlang
rlang::quoandrlang::quosmatchrlang::expr()andrlang::exprs()rlang::eval_tidy()evaluates quosures
foo <- function(x) enquo(x)
foo(a + b)## <quosure>
## expr: ^a + b
## env: globalq1 <- new_quosure(expr(x + y), env(x = 1, y = 10))
eval_tidy(q1)## [1] 11Data Masks
Call variables from a data frame using expressions. eval_tidy() excepts a dataframe in addition to the environment in the closure which it unpacks and makes available to the evaluation of the expression.
eval() excepts a dataframe as an environment
with2 <- function(data, expr) {
expr <- enquo(expr)
eval_tidy(expr, data = data)
}
df <- data.frame(y = 1:10)
x <- 100
with2(df, x * y)## [1] 100 200 300 400 500 600 700 800 900 1000with3 <- function(data, expr) {
expr <- substitute(expr)
eval(expr, envir = data)
}
with3(df, x * y)## [1] 100 200 300 400 500 600 700 800 900 1000for rlang quosures we can solve ambiguity between the dataframe and the environment using .data and .env
df <- data.frame(y = 1:10, x = 11:20)
x <- 100
with2(df, .data$x + .data$y)## [1] 12 14 16 18 20 22 24 26 28 30with2(df, .env$x + .data$y)## [1] 101 102 103 104 105 106 107 108 109 110# does not work with the base version
with3(df, .data$x)## NULLBase Function Using Datamasks
subset
similar to dplyr::filter
subset2 <- function(df, expr) {
qu <- enquo(expr)
bool <- eval_tidy(qu, df)
browser
stopifnot(is.logical(bool))
df[bool,]
}
subset2(mtcars, cyl == 6)## mpg cyl disp hp drat wt qsec vs am gear carb
## Mazda RX4 21.0 6 160.0 110 3.90 2.620 16.46 0 1 4 4
## Mazda RX4 Wag 21.0 6 160.0 110 3.90 2.875 17.02 0 1 4 4
## Hornet 4 Drive 21.4 6 258.0 110 3.08 3.215 19.44 1 0 3 1
## Valiant 18.1 6 225.0 105 2.76 3.460 20.22 1 0 3 1
## Merc 280 19.2 6 167.6 123 3.92 3.440 18.30 1 0 4 4
## Merc 280C 17.8 6 167.6 123 3.92 3.440 18.90 1 0 4 4
## Ferrari Dino 19.7 6 145.0 175 3.62 2.770 15.50 0 1 5 6transform
similar to dplyr::mutate
transform2 <- function(df, ...) {
dots <- enquos(...)
for (i in seq_along(dots)) {
col <- names(dots)[i]
df[[col]] <- eval_tidy(dots[[i]], df)
}
return(df)
}
df <- data.frame(x = 1:10, y = 11: 10)
transform2(df, z = x * y, z0 = z + 1)## x y z z0
## 1 1 11 11 12
## 2 2 10 20 21
## 3 3 11 33 34
## 4 4 10 40 41
## 5 5 11 55 56
## 6 6 10 60 61
## 7 7 11 77 78
## 8 8 10 80 81
## 9 9 11 99 100
## 10 10 10 100 101select
rewrite of dplyr::select()
select2 <- function(data, ...) {
dots <- enquos(...)
vars <- as.list(set_names(seq_along(data), names(data)))
cols <- unlist(map(dots, eval_tidy, vars))
data[, cols, drop = FALSE]
}
df <- data.frame(a = 1, b = 2, c = 3, d = 4, e = 5)
select2(df, b:d, a)## b c d a
## 1 2 3 4 1Programming with rlang/dplyr
Pass expressions onto functions using expression
We cannot pass them directly. We need to quote and selectively unquote
We can use quosures and bangs
sample_col <- function(df, col, n = 5) {
qu_col <- enquo(col)
head(select2(df, !! qu_col), n)
}
sample_col(mtcars, disp)## disp
## Mazda RX4 160
## Mazda RX4 Wag 160
## Datsun 710 108
## Hornet 4 Drive 258
## Hornet Sportabout 360sample_cols <- function(df, ..., n = 5) {
qu_cols <- enquos(...)
head(select2(df, !!! qu_cols), n)
}
sample_cols(mtcars, disp, cyl)## disp cyl
## Mazda RX4 160 6
## Mazda RX4 Wag 160 6
## Datsun 710 108 4
## Hornet 4 Drive 258 6
## Hornet Sportabout 360 8Or expressions
sample_col <- function(df, col, n = 5) {
qu_col <- enexpr(col)
head(select(df, !! qu_col), n)
}
sample_col(mtcars, disp)## disp
## Mazda RX4 160
## Mazda RX4 Wag 160
## Datsun 710 108
## Hornet 4 Drive 258
## Hornet Sportabout 360sample_cols <- function(df, ..., n = 5) {
qu_cols <- enexprs(...)
head(select2(df, !!! qu_cols), n)
}
sample_cols(mtcars, disp, cyl)## disp cyl
## Mazda RX4 160 6
## Mazda RX4 Wag 160 6
## Datsun 710 108 4
## Hornet 4 Drive 258 6
## Hornet Sportabout 360 8Or Curly Curl
**new standard as of rlang 0.4.0
sample_col <- function(df, col, n = 5) {
head(select2(df, {{col}}), n)
}
sample_col(mtcars, disp)## disp
## Mazda RX4 160
## Mazda RX4 Wag 160
## Datsun 710 108
## Hornet 4 Drive 258
## Hornet Sportabout 360sample_cols <- function(df, ..., n = 5) {
head(select2(df, ...), n)
}
sample_cols(mtcars, disp, cyl)## disp cyl
## Mazda RX4 160 6
## Mazda RX4 Wag 160 6
## Datsun 710 108 4
## Hornet 4 Drive 258 6
## Hornet Sportabout 360 8Pass strings onto functions using expression
use .data
Applications
Fetching Model Training Data from Global Environment
save the call using match.call()
slope_model <- function(data, form) {
m <- lm(form, data)
structure(
list(
slope = m$coefficients[[2]],
intercept = m$coefficients[[1]],
call = match.call()
),
class = "slope_model"
)
}
plot.slope_model <- function(m) {
stopifnot(inherits(m, "slope_model"))
data_expr <- m$call[["data"]]
stopifnot(exists(deparse(data_expr)))
cols <- colnames(eval(data_expr))
form <- m$call[["form"]]
y_expr <- rlang::f_lhs(form)
x_expr <- rlang::f_rhs(form)
y_str <- deparse(y_expr)
x_str <- deparse(x_expr)
stopifnot(c(y_str, y_str) %in% cols)
eval(data_expr) %>%
select({{y_expr}}, {{x_expr}}) %>%
ggplot(aes({{x_expr}}, {{y_expr}})) +
geom_point() +
geom_abline(slope = m$slope, intercept = m$intercept) +
theme_minimal()
}
m <- slope_model(mtcars, disp ~ hp)
m## $slope
## [1] 1.42977
##
## $intercept
## [1] 20.99248
##
## $call
## slope_model(data = mtcars, form = disp ~ hp)
##
## attr(,"class")
## [1] "slope_model"plot(m)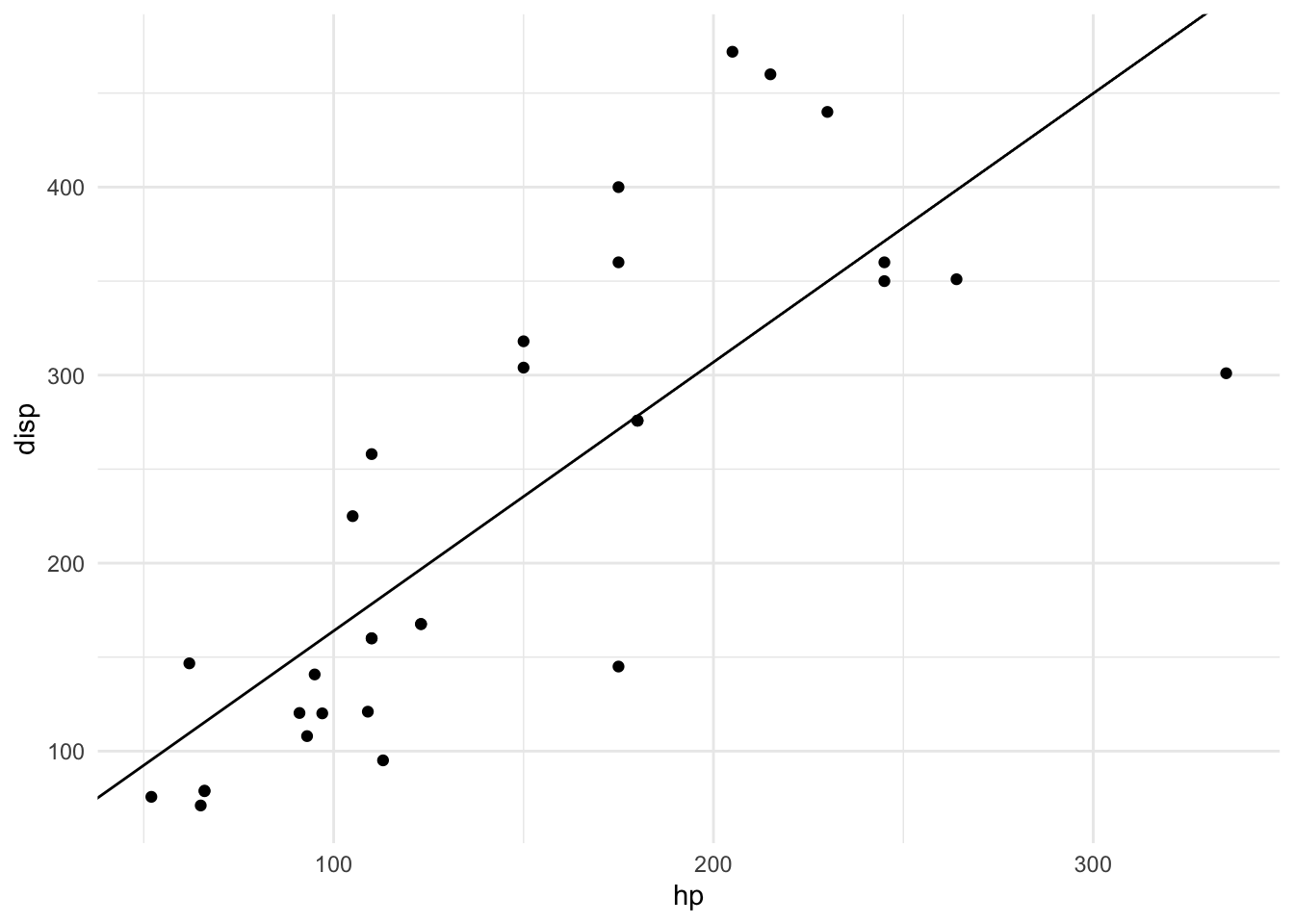
pryr::object_size(m)## 1,520 Bpryr::object_size({m$data <- mtcars})## 7,208 BCorrecting Calls for Wrapped Modelling Functions
when we write a wrapper the saved call cannot be used to reconstruct the actual call. And methods relying on it will not work
wr_slope <- function(data, form) {
slope_model(data, form)
}
m <- wr_slope(mtcars, disp ~ hp)
m## $slope
## [1] 1.42977
##
## $intercept
## [1] 20.99248
##
## $call
## slope_model(data = data, form = form)
##
## attr(,"class")
## [1] "slope_model"plot(m)## Error: `x` must be a formulawe need to reconstruct a new call inside the wrapper and pass it to the modelling function.
wr_slope <- function(data, form) {
data <- enexpr(data)
form <- enexpr(form)
new_call <- expr(slope_model(!!data, !!form))
eval(new_call)
}
m <- wr_slope(mtcars, disp ~ hp)
m## $slope
## [1] 1.42977
##
## $intercept
## [1] 20.99248
##
## $call
## slope_model(data = mtcars, form = disp ~ hp)
##
## attr(,"class")
## [1] "slope_model"plot(m)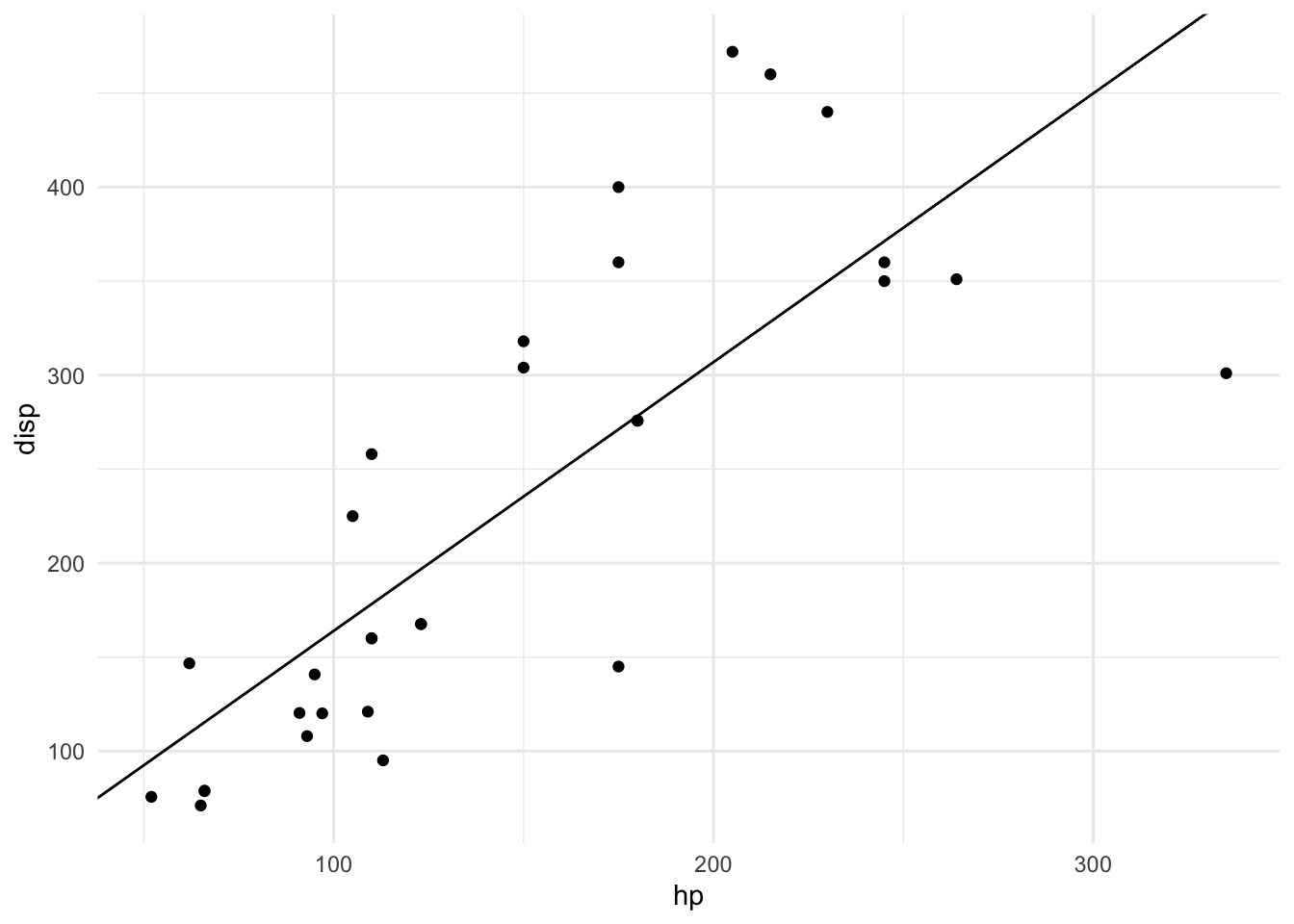
attaching the call to the model object is risky because if the modeling function is used by do.call or purr::map we risk attaching the entire data to the call.
do.call(slope_model, list(mtcars, disp ~ hp))## $slope
## [1] 1.42977
##
## $intercept
## [1] 20.99248
##
## $call
## (function(data, form) {
## m <- lm(form, data)
## structure(
## list(
## slope = m$coefficients[[2]],
## intercept = m$coefficients[[1]],
## call = match.call()
## ),
## class = "slope_model"
## )
## })(data = list(mpg = c(21, 21, 22.8, 21.4, 18.7, 18.1, 14.3,
## 24.4, 22.8, 19.2, 17.8, 16.4, 17.3, 15.2, 10.4, 10.4, 14.7, 32.4,
## 30.4, 33.9, 21.5, 15.5, 15.2, 13.3, 19.2, 27.3, 26, 30.4, 15.8,
## 19.7, 15, 21.4), cyl = c(6, 6, 4, 6, 8, 6, 8, 4, 4, 6, 6, 8,
## 8, 8, 8, 8, 8, 4, 4, 4, 4, 8, 8, 8, 8, 4, 4, 4, 8, 6, 8, 4),
## disp = c(160, 160, 108, 258, 360, 225, 360, 146.7, 140.8,
## 167.6, 167.6, 275.8, 275.8, 275.8, 472, 460, 440, 78.7, 75.7,
## 71.1, 120.1, 318, 304, 350, 400, 79, 120.3, 95.1, 351, 145,
## 301, 121), hp = c(110, 110, 93, 110, 175, 105, 245, 62, 95,
## 123, 123, 180, 180, 180, 205, 215, 230, 66, 52, 65, 97, 150,
## 150, 245, 175, 66, 91, 113, 264, 175, 335, 109), drat = c(3.9,
## 3.9, 3.85, 3.08, 3.15, 2.76, 3.21, 3.69, 3.92, 3.92, 3.92,
## 3.07, 3.07, 3.07, 2.93, 3, 3.23, 4.08, 4.93, 4.22, 3.7, 2.76,
## 3.15, 3.73, 3.08, 4.08, 4.43, 3.77, 4.22, 3.62, 3.54, 4.11
## ), wt = c(2.62, 2.875, 2.32, 3.215, 3.44, 3.46, 3.57, 3.19,
## 3.15, 3.44, 3.44, 4.07, 3.73, 3.78, 5.25, 5.424, 5.345, 2.2,
## 1.615, 1.835, 2.465, 3.52, 3.435, 3.84, 3.845, 1.935, 2.14,
## 1.513, 3.17, 2.77, 3.57, 2.78), qsec = c(16.46, 17.02, 18.61,
## 19.44, 17.02, 20.22, 15.84, 20, 22.9, 18.3, 18.9, 17.4, 17.6,
## 18, 17.98, 17.82, 17.42, 19.47, 18.52, 19.9, 20.01, 16.87,
## 17.3, 15.41, 17.05, 18.9, 16.7, 16.9, 14.5, 15.5, 14.6, 18.6
## ), vs = c(0, 0, 1, 1, 0, 1, 0, 1, 1, 1, 1, 0, 0, 0, 0, 0,
## 0, 1, 1, 1, 1, 0, 0, 0, 0, 1, 0, 1, 0, 0, 0, 1), am = c(1,
## 1, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 1, 1,
## 0, 0, 0, 0, 0, 1, 1, 1, 1, 1, 1, 1), gear = c(4, 4, 4, 3,
## 3, 3, 3, 4, 4, 4, 4, 3, 3, 3, 3, 3, 3, 4, 4, 4, 3, 3, 3,
## 3, 3, 4, 5, 5, 5, 5, 5, 4), carb = c(4, 4, 1, 1, 2, 1, 4,
## 2, 2, 4, 4, 3, 3, 3, 4, 4, 4, 1, 2, 1, 1, 2, 2, 4, 2, 1,
## 2, 2, 4, 6, 8, 2)), form = disp ~ hp)
##
## attr(,"class")
## [1] "slope_model"do.call(wr_slope, list(mtcars, disp ~ hp))## $slope
## [1] 1.42977
##
## $intercept
## [1] 20.99248
##
## $call
## slope_model(data = list(mpg = c(21, 21, 22.8, 21.4, 18.7, 18.1,
## 14.3, 24.4, 22.8, 19.2, 17.8, 16.4, 17.3, 15.2, 10.4, 10.4, 14.7,
## 32.4, 30.4, 33.9, 21.5, 15.5, 15.2, 13.3, 19.2, 27.3, 26, 30.4,
## 15.8, 19.7, 15, 21.4), cyl = c(6, 6, 4, 6, 8, 6, 8, 4, 4, 6,
## 6, 8, 8, 8, 8, 8, 8, 4, 4, 4, 4, 8, 8, 8, 8, 4, 4, 4, 8, 6, 8,
## 4), disp = c(160, 160, 108, 258, 360, 225, 360, 146.7, 140.8,
## 167.6, 167.6, 275.8, 275.8, 275.8, 472, 460, 440, 78.7, 75.7,
## 71.1, 120.1, 318, 304, 350, 400, 79, 120.3, 95.1, 351, 145, 301,
## 121), hp = c(110, 110, 93, 110, 175, 105, 245, 62, 95, 123, 123,
## 180, 180, 180, 205, 215, 230, 66, 52, 65, 97, 150, 150, 245,
## 175, 66, 91, 113, 264, 175, 335, 109), drat = c(3.9, 3.9, 3.85,
## 3.08, 3.15, 2.76, 3.21, 3.69, 3.92, 3.92, 3.92, 3.07, 3.07, 3.07,
## 2.93, 3, 3.23, 4.08, 4.93, 4.22, 3.7, 2.76, 3.15, 3.73, 3.08,
## 4.08, 4.43, 3.77, 4.22, 3.62, 3.54, 4.11), wt = c(2.62, 2.875,
## 2.32, 3.215, 3.44, 3.46, 3.57, 3.19, 3.15, 3.44, 3.44, 4.07,
## 3.73, 3.78, 5.25, 5.424, 5.345, 2.2, 1.615, 1.835, 2.465, 3.52,
## 3.435, 3.84, 3.845, 1.935, 2.14, 1.513, 3.17, 2.77, 3.57, 2.78
## ), qsec = c(16.46, 17.02, 18.61, 19.44, 17.02, 20.22, 15.84,
## 20, 22.9, 18.3, 18.9, 17.4, 17.6, 18, 17.98, 17.82, 17.42, 19.47,
## 18.52, 19.9, 20.01, 16.87, 17.3, 15.41, 17.05, 18.9, 16.7, 16.9,
## 14.5, 15.5, 14.6, 18.6), vs = c(0, 0, 1, 1, 0, 1, 0, 1, 1, 1,
## 1, 0, 0, 0, 0, 0, 0, 1, 1, 1, 1, 0, 0, 0, 0, 1, 0, 1, 0, 0, 0,
## 1), am = c(1, 1, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
## 1, 1, 1, 0, 0, 0, 0, 0, 1, 1, 1, 1, 1, 1, 1), gear = c(4, 4,
## 4, 3, 3, 3, 3, 4, 4, 4, 4, 3, 3, 3, 3, 3, 3, 4, 4, 4, 3, 3, 3,
## 3, 3, 4, 5, 5, 5, 5, 5, 4), carb = c(4, 4, 1, 1, 2, 1, 4, 2,
## 2, 4, 4, 3, 3, 3, 4, 4, 4, 1, 2, 1, 1, 2, 2, 4, 2, 1, 2, 2, 4,
## 6, 8, 2)), form = disp ~ hp)
##
## attr(,"class")
## [1] "slope_model"do.call(lm, list(disp ~ hp, mtcars))##
## Call:
## (function (formula, data, subset, weights, na.action, method = "qr",
## model = TRUE, x = FALSE, y = FALSE, qr = TRUE, singular.ok = TRUE,
## contrasts = NULL, offset, ...)
## {
## ret.x <- x
## ret.y <- y
## cl <- match.call()
## mf <- match.call(expand.dots = FALSE)
## m <- match(c("formula", "data", "subset", "weights", "na.action",
## "offset"), names(mf), 0L)
## mf <- mf[c(1L, m)]
## mf$drop.unused.levels <- TRUE
## mf[[1L]] <- quote(stats::model.frame)
## mf <- eval(mf, parent.frame())
## if (method == "model.frame")
## return(mf)
## else if (method != "qr")
## warning(gettextf("method = '%s' is not supported. Using 'qr'",
## method), domain = NA)
## mt <- attr(mf, "terms")
## y <- model.response(mf, "numeric")
## w <- as.vector(model.weights(mf))
## if (!is.null(w) && !is.numeric(w))
## stop("'weights' must be a numeric vector")
## offset <- model.offset(mf)
## mlm <- is.matrix(y)
## ny <- if (mlm)
## nrow(y)
## else length(y)
## if (!is.null(offset)) {
## if (!mlm)
## offset <- as.vector(offset)
## if (NROW(offset) != ny)
## stop(gettextf("number of offsets is %d, should equal %d (number of observations)",
## NROW(offset), ny), domain = NA)
## }
## if (is.empty.model(mt)) {
## x <- NULL
## z <- list(coefficients = if (mlm) matrix(NA_real_, 0,
## ncol(y)) else numeric(), residuals = y, fitted.values = 0 *
## y, weights = w, rank = 0L, df.residual = if (!is.null(w)) sum(w !=
## 0) else ny)
## if (!is.null(offset)) {
## z$fitted.values <- offset
## z$residuals <- y - offset
## }
## }
## else {
## x <- model.matrix(mt, mf, contrasts)
## z <- if (is.null(w))
## lm.fit(x, y, offset = offset, singular.ok = singular.ok,
## ...)
## else lm.wfit(x, y, w, offset = offset, singular.ok = singular.ok,
## ...)
## }
## class(z) <- c(if (mlm) "mlm", "lm")
## z$na.action <- attr(mf, "na.action")
## z$offset <- offset
## z$contrasts <- attr(x, "contrasts")
## z$xlevels <- .getXlevels(mt, mf)
## z$call <- cl
## z$terms <- mt
## if (model)
## z$model <- mf
## if (ret.x)
## z$x <- x
## if (ret.y)
## z$y <- y
## if (!qr)
## z$qr <- NULL
## z
## })(formula = disp ~ hp, data = structure(list(mpg = c(21, 21,
## 22.8, 21.4, 18.7, 18.1, 14.3, 24.4, 22.8, 19.2, 17.8, 16.4, 17.3,
## 15.2, 10.4, 10.4, 14.7, 32.4, 30.4, 33.9, 21.5, 15.5, 15.2, 13.3,
## 19.2, 27.3, 26, 30.4, 15.8, 19.7, 15, 21.4), cyl = c(6, 6, 4,
## 6, 8, 6, 8, 4, 4, 6, 6, 8, 8, 8, 8, 8, 8, 4, 4, 4, 4, 8, 8, 8,
## 8, 4, 4, 4, 8, 6, 8, 4), disp = c(160, 160, 108, 258, 360, 225,
## 360, 146.7, 140.8, 167.6, 167.6, 275.8, 275.8, 275.8, 472, 460,
## 440, 78.7, 75.7, 71.1, 120.1, 318, 304, 350, 400, 79, 120.3,
## 95.1, 351, 145, 301, 121), hp = c(110, 110, 93, 110, 175, 105,
## 245, 62, 95, 123, 123, 180, 180, 180, 205, 215, 230, 66, 52,
## 65, 97, 150, 150, 245, 175, 66, 91, 113, 264, 175, 335, 109),
## drat = c(3.9, 3.9, 3.85, 3.08, 3.15, 2.76, 3.21, 3.69, 3.92,
## 3.92, 3.92, 3.07, 3.07, 3.07, 2.93, 3, 3.23, 4.08, 4.93,
## 4.22, 3.7, 2.76, 3.15, 3.73, 3.08, 4.08, 4.43, 3.77, 4.22,
## 3.62, 3.54, 4.11), wt = c(2.62, 2.875, 2.32, 3.215, 3.44,
## 3.46, 3.57, 3.19, 3.15, 3.44, 3.44, 4.07, 3.73, 3.78, 5.25,
## 5.424, 5.345, 2.2, 1.615, 1.835, 2.465, 3.52, 3.435, 3.84,
## 3.845, 1.935, 2.14, 1.513, 3.17, 2.77, 3.57, 2.78), qsec = c(16.46,
## 17.02, 18.61, 19.44, 17.02, 20.22, 15.84, 20, 22.9, 18.3,
## 18.9, 17.4, 17.6, 18, 17.98, 17.82, 17.42, 19.47, 18.52,
## 19.9, 20.01, 16.87, 17.3, 15.41, 17.05, 18.9, 16.7, 16.9,
## 14.5, 15.5, 14.6, 18.6), vs = c(0, 0, 1, 1, 0, 1, 0, 1, 1,
## 1, 1, 0, 0, 0, 0, 0, 0, 1, 1, 1, 1, 0, 0, 0, 0, 1, 0, 1,
## 0, 0, 0, 1), am = c(1, 1, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
## 0, 0, 0, 0, 1, 1, 1, 0, 0, 0, 0, 0, 1, 1, 1, 1, 1, 1, 1),
## gear = c(4, 4, 4, 3, 3, 3, 3, 4, 4, 4, 4, 3, 3, 3, 3, 3,
## 3, 4, 4, 4, 3, 3, 3, 3, 3, 4, 5, 5, 5, 5, 5, 4), carb = c(4,
## 4, 1, 1, 2, 1, 4, 2, 2, 4, 4, 3, 3, 3, 4, 4, 4, 1, 2, 1,
## 1, 2, 2, 4, 2, 1, 2, 2, 4, 6, 8, 2)), row.names = c("Mazda RX4",
## "Mazda RX4 Wag", "Datsun 710", "Hornet 4 Drive", "Hornet Sportabout",
## "Valiant", "Duster 360", "Merc 240D", "Merc 230", "Merc 280",
## "Merc 280C", "Merc 450SE", "Merc 450SL", "Merc 450SLC", "Cadillac Fleetwood",
## "Lincoln Continental", "Chrysler Imperial", "Fiat 128", "Honda Civic",
## "Toyota Corolla", "Toyota Corona", "Dodge Challenger", "AMC Javelin",
## "Camaro Z28", "Pontiac Firebird", "Fiat X1-9", "Porsche 914-2",
## "Lotus Europa", "Ford Pantera L", "Ferrari Dino", "Maserati Bora",
## "Volvo 142E"), class = "data.frame"))
##
## Coefficients:
## (Intercept) hp
## 20.99 1.43This is why a size check for the saving the call is required in the tidymodels convention guide
df_m <- mtcars %>%
group_by(cyl) %>%
nest() %>%
mutate(m = map(data, wr_slope, disp ~ hp))
map(df_m$m, ~ .$call)## [[1]]
## slope_model(data = list(mpg = c(21, 21, 21.4, 18.1, 19.2, 17.8,
## 19.7), disp = c(160, 160, 258, 225, 167.6, 167.6, 145), hp = c(110,
## 110, 110, 105, 123, 123, 175), drat = c(3.9, 3.9, 3.08, 2.76,
## 3.92, 3.92, 3.62), wt = c(2.62, 2.875, 3.215, 3.46, 3.44, 3.44,
## 2.77), qsec = c(16.46, 17.02, 19.44, 20.22, 18.3, 18.9, 15.5),
## vs = c(0, 0, 1, 1, 1, 1, 0), am = c(1, 1, 0, 0, 0, 0, 1),
## gear = c(4, 4, 3, 3, 4, 4, 5), carb = c(4, 4, 1, 1, 4, 4,
## 6)), form = disp ~ hp)
##
## [[2]]
## slope_model(data = list(mpg = c(22.8, 24.4, 22.8, 32.4, 30.4,
## 33.9, 21.5, 27.3, 26, 30.4, 21.4), disp = c(108, 146.7, 140.8,
## 78.7, 75.7, 71.1, 120.1, 79, 120.3, 95.1, 121), hp = c(93, 62,
## 95, 66, 52, 65, 97, 66, 91, 113, 109), drat = c(3.85, 3.69, 3.92,
## 4.08, 4.93, 4.22, 3.7, 4.08, 4.43, 3.77, 4.11), wt = c(2.32,
## 3.19, 3.15, 2.2, 1.615, 1.835, 2.465, 1.935, 2.14, 1.513, 2.78
## ), qsec = c(18.61, 20, 22.9, 19.47, 18.52, 19.9, 20.01, 18.9,
## 16.7, 16.9, 18.6), vs = c(1, 1, 1, 1, 1, 1, 1, 1, 0, 1, 1), am = c(1,
## 0, 0, 1, 1, 1, 0, 1, 1, 1, 1), gear = c(4, 4, 4, 4, 4, 4, 3,
## 4, 5, 5, 4), carb = c(1, 2, 2, 1, 2, 1, 1, 1, 2, 2, 2)), form = disp ~
## hp)
##
## [[3]]
## slope_model(data = list(mpg = c(18.7, 14.3, 16.4, 17.3, 15.2,
## 10.4, 10.4, 14.7, 15.5, 15.2, 13.3, 19.2, 15.8, 15), disp = c(360,
## 360, 275.8, 275.8, 275.8, 472, 460, 440, 318, 304, 350, 400,
## 351, 301), hp = c(175, 245, 180, 180, 180, 205, 215, 230, 150,
## 150, 245, 175, 264, 335), drat = c(3.15, 3.21, 3.07, 3.07, 3.07,
## 2.93, 3, 3.23, 2.76, 3.15, 3.73, 3.08, 4.22, 3.54), wt = c(3.44,
## 3.57, 4.07, 3.73, 3.78, 5.25, 5.424, 5.345, 3.52, 3.435, 3.84,
## 3.845, 3.17, 3.57), qsec = c(17.02, 15.84, 17.4, 17.6, 18, 17.98,
## 17.82, 17.42, 16.87, 17.3, 15.41, 17.05, 14.5, 14.6), vs = c(0,
## 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0), am = c(0, 0, 0, 0, 0,
## 0, 0, 0, 0, 0, 0, 0, 1, 1), gear = c(3, 3, 3, 3, 3, 3, 3, 3,
## 3, 3, 3, 3, 5, 5), carb = c(2, 4, 3, 3, 3, 4, 4, 4, 2, 2, 4,
## 2, 4, 8)), form = disp ~ hp)map(df_m$m, plot)## Error in exists(deparse(data_expr)): first argument has length > 1instead attach arguments individually
training_data <- function(name, df) {
structure(
list(
name = stringr::str_trunc(name, width = 120)[[1]],
cols = colnames(df),
dim = dim(df)
),
class = "train_data"
)
}
match_training_data <- function(df, train_data) {
stopifnot(is.data.frame(df))
stopifnot(all(train_data$cols %in% colnames(df)))
df <- df[, train_data$cols]
stopifnot(all(dim(df) == train_data$dim))
return(TRUE)
}
slope_model2 <- function(data, form) {
y <- deparse(rlang::f_lhs(form))
x <- deparse(rlang::f_rhs(form))
data_name <- deparse(rlang::enexpr(data))
train_data <- training_data(data_name, data[, c(x, y)])
m <- lm(form, data)
structure(
list(
slope = m$coefficients[[2]],
intercept = m$coefficients[[1]],
train_data = train_data,
x = x,
y = y
),
class = "slope_model"
)
}
plot.slope_model <- function(m, data = NULL) {
stopifnot(inherits(m, "slope_model"))
if (is.null(data) & exists(m$train_data$name)) {
df_train <- eval(parse(text = m$train_data$name))
match_training_data(df_train, m$train_data)
} else if (! is.null(data)) {
df_train <- data
match_training_data(df_train, m$train_data)
} else {
stop("training data not found")
}
ggplot(df_train, aes_string(m$x, m$y)) +
geom_point() +
geom_abline(slope = m$slope, intercept = m$intercept) +
theme_minimal()
}
m <- slope_model2(mtcars, disp ~ hp)
str(m)## List of 5
## $ slope : num 1.43
## $ intercept : num 21
## $ train_data:List of 3
## ..$ name: chr "mtcars"
## ..$ cols: chr [1:2] "hp" "disp"
## ..$ dim : int [1:2] 32 2
## ..- attr(*, "class")= chr "train_data"
## $ x : chr "hp"
## $ y : chr "disp"
## - attr(*, "class")= chr "slope_model"plot(m)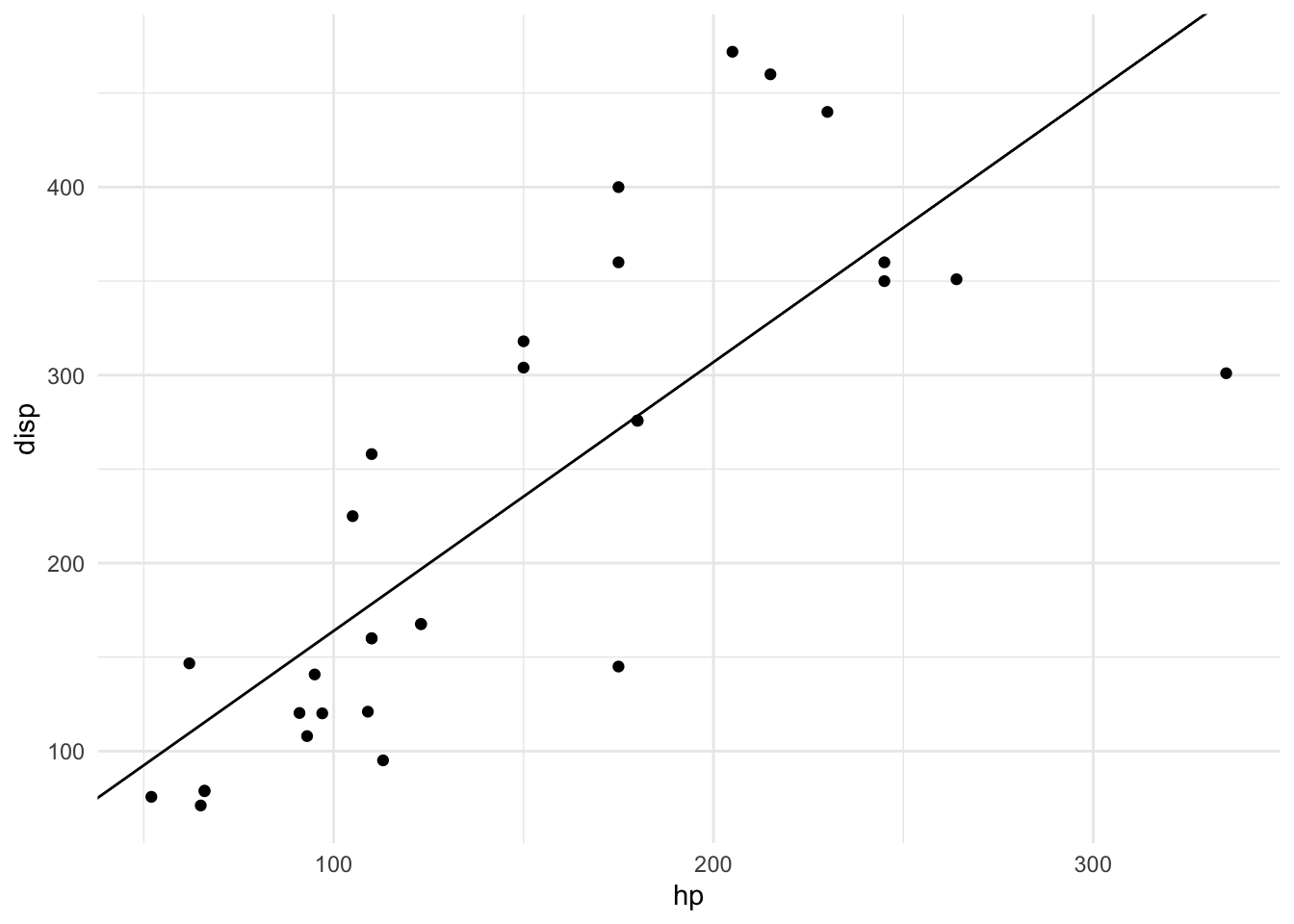
check wrapper compatibility
wr_slope <- function(data, form) {
slope_model2(data, form)
}
m <- wr_slope(mtcars, disp ~ hp)
str(m)## List of 5
## $ slope : num 1.43
## $ intercept : num 21
## $ train_data:List of 3
## ..$ name: chr "data"
## ..$ cols: chr [1:2] "hp" "disp"
## ..$ dim : int [1:2] 32 2
## ..- attr(*, "class")= chr "train_data"
## $ x : chr "hp"
## $ y : chr "disp"
## - attr(*, "class")= chr "slope_model"plot(m)## Error in match_training_data(df_train, m$train_data): is.data.frame(df) is not TRUEplot(m, mtcars)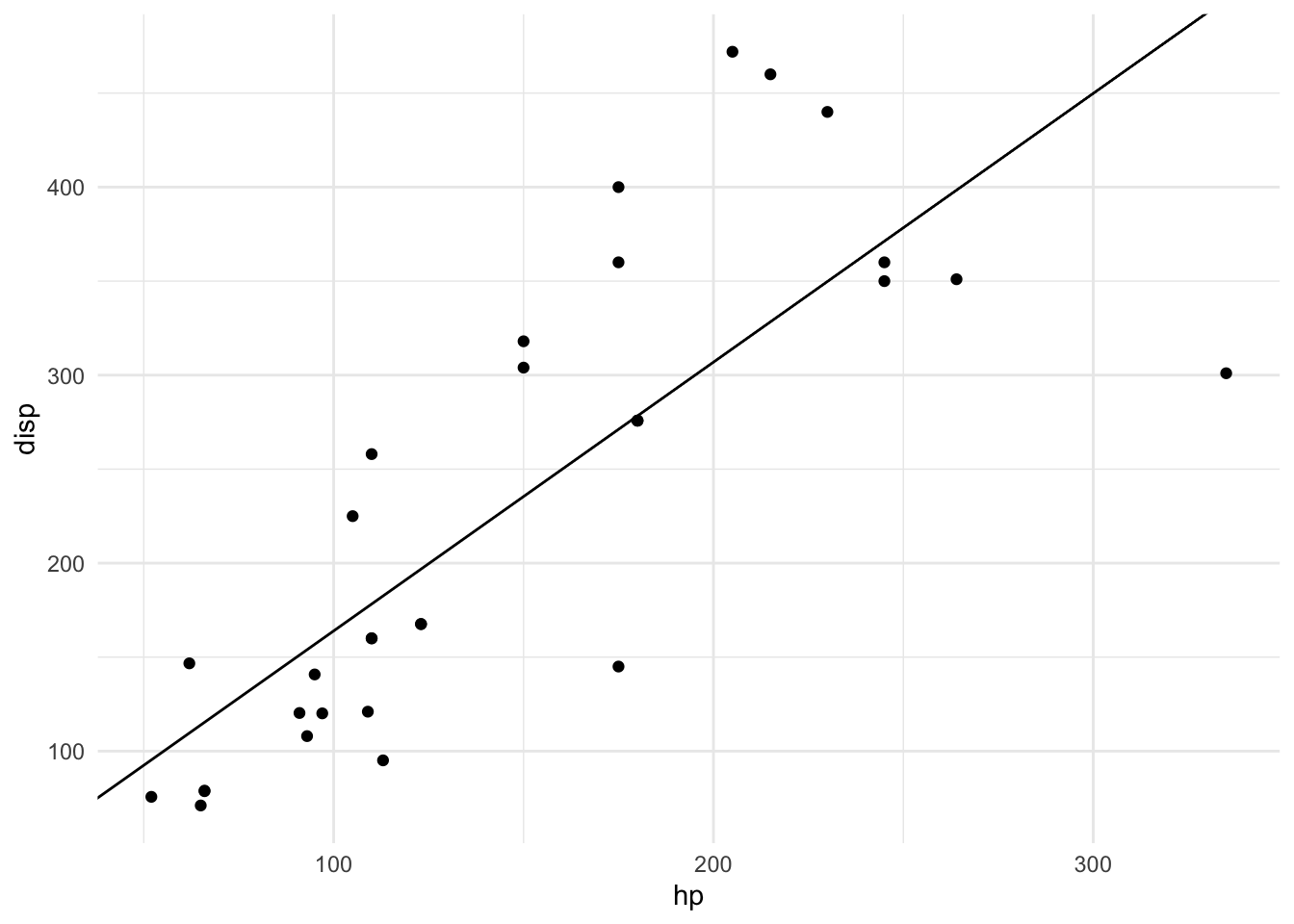
still need it
wr_slope2 <- function(data, form) {
data <- enexpr(data)
form <- enexpr(form)
new_call <- expr(slope_model2(!!data, !!form))
eval(new_call)
}
m <- wr_slope2(mtcars, disp ~ hp)
str(m)## List of 5
## $ slope : num 1.43
## $ intercept : num 21
## $ train_data:List of 3
## ..$ name: chr "mtcars"
## ..$ cols: chr [1:2] "hp" "disp"
## ..$ dim : int [1:2] 32 2
## ..- attr(*, "class")= chr "train_data"
## $ x : chr "hp"
## $ y : chr "disp"
## - attr(*, "class")= chr "slope_model"plot(m)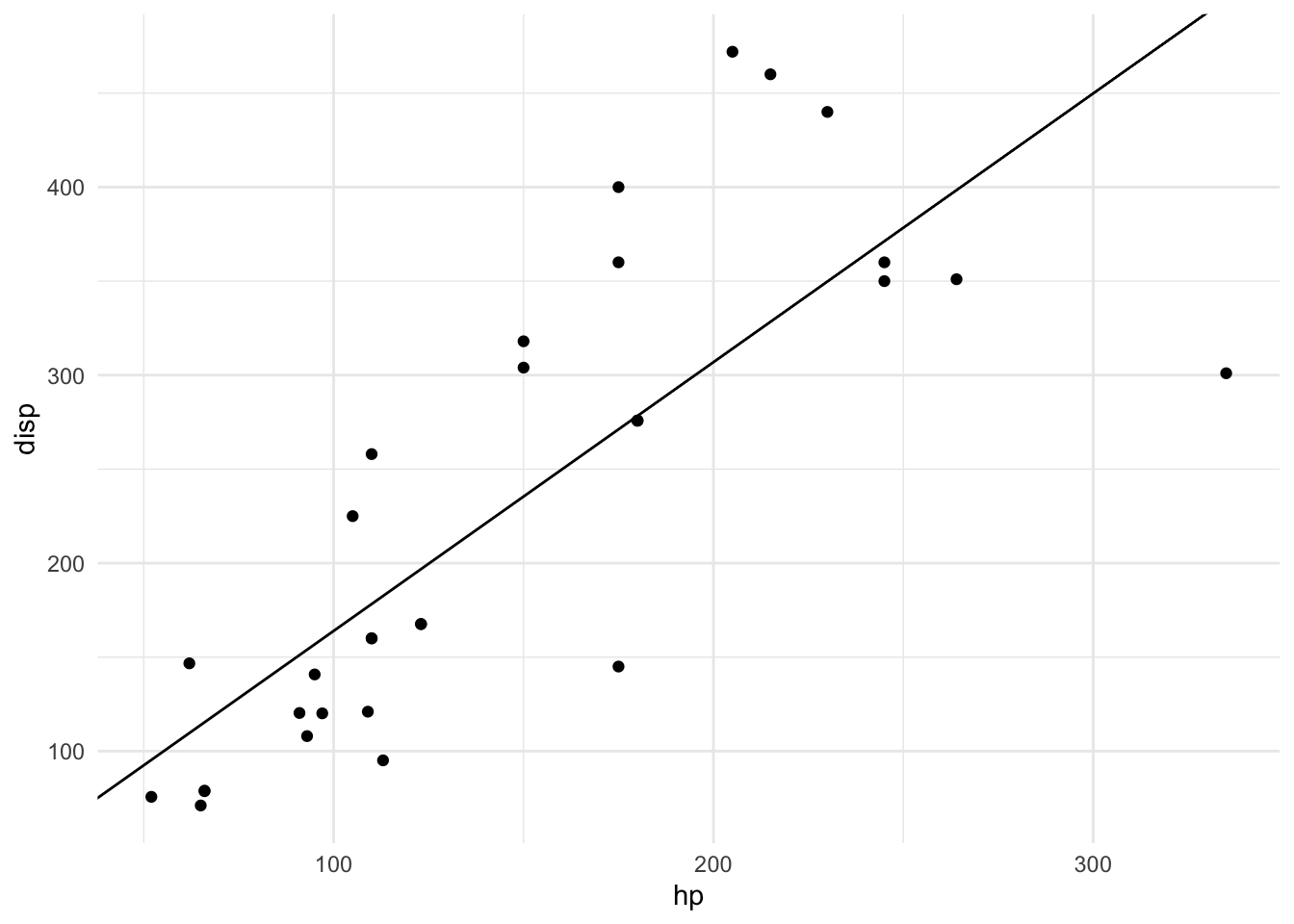
do.call
m <- do.call(slope_model2, list(mtcars, disp ~ hp))
str(m)## List of 5
## $ slope : num 1.43
## $ intercept : num 21
## $ train_data:List of 3
## ..$ name: chr "structure(list(mpg = c(21, 21, 22.8, 21.4, 18.7, 18.1, 14.3, "
## ..$ cols: chr [1:2] "hp" "disp"
## ..$ dim : int [1:2] 32 2
## ..- attr(*, "class")= chr "train_data"
## $ x : chr "hp"
## $ y : chr "disp"
## - attr(*, "class")= chr "slope_model"plot(m)## Error in plot.slope_model(m): training data not foundplot(m, mtcars)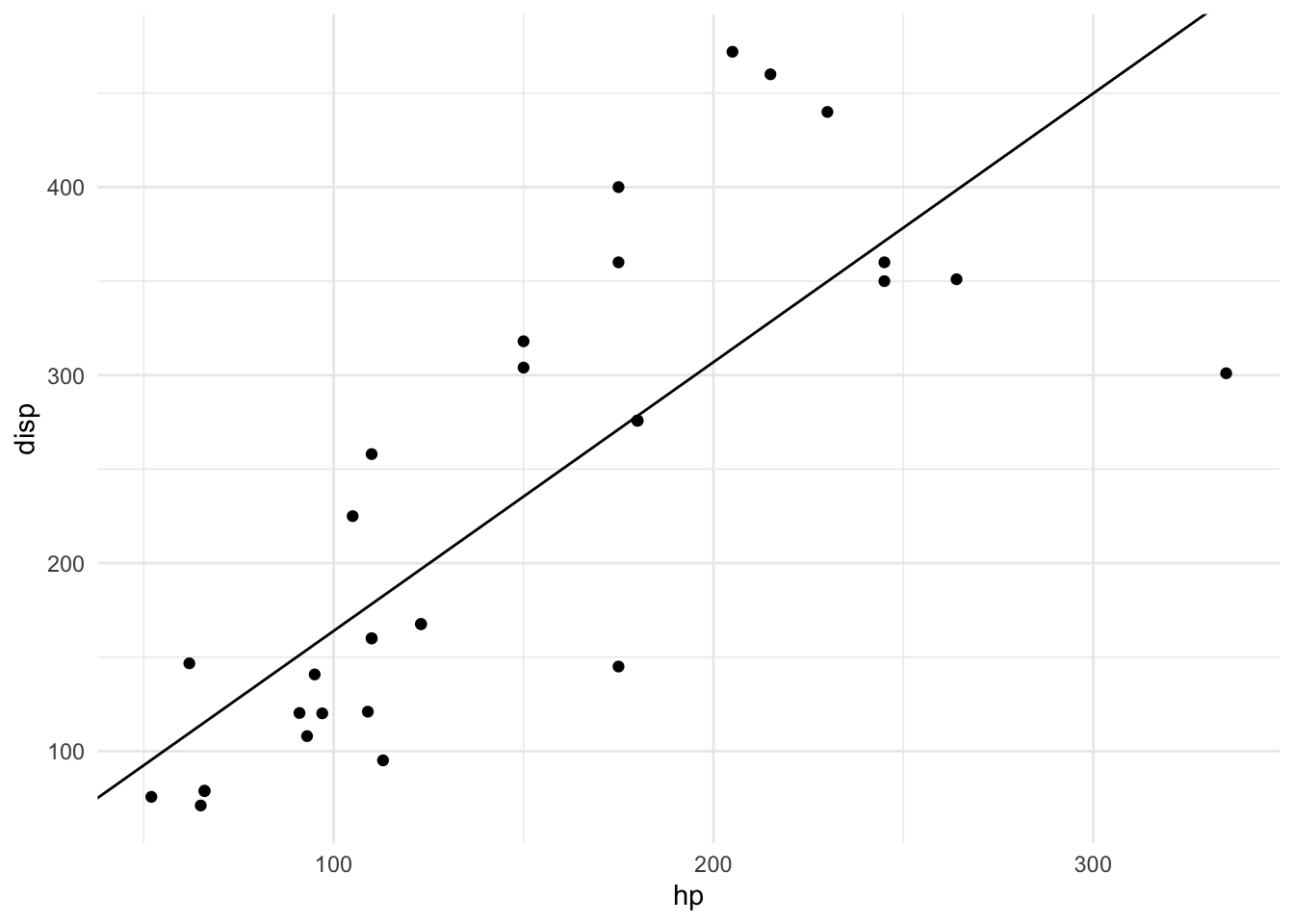
Printing R Code for ggplots Plots
we want to display the r code necessary to render a plot
plot_vars <- function(df, x, y, smooth = FALSE) {
df <- enexpr(df)
x <- enexpr(x)
y <- enexpr(y)
ex <- expr(
ggplot(!!df, aes(!! x, !!y)) +
geom_point()
)
if(smooth){
ex <- expr(!! ex + geom_smooth())
}
return(ex)
}
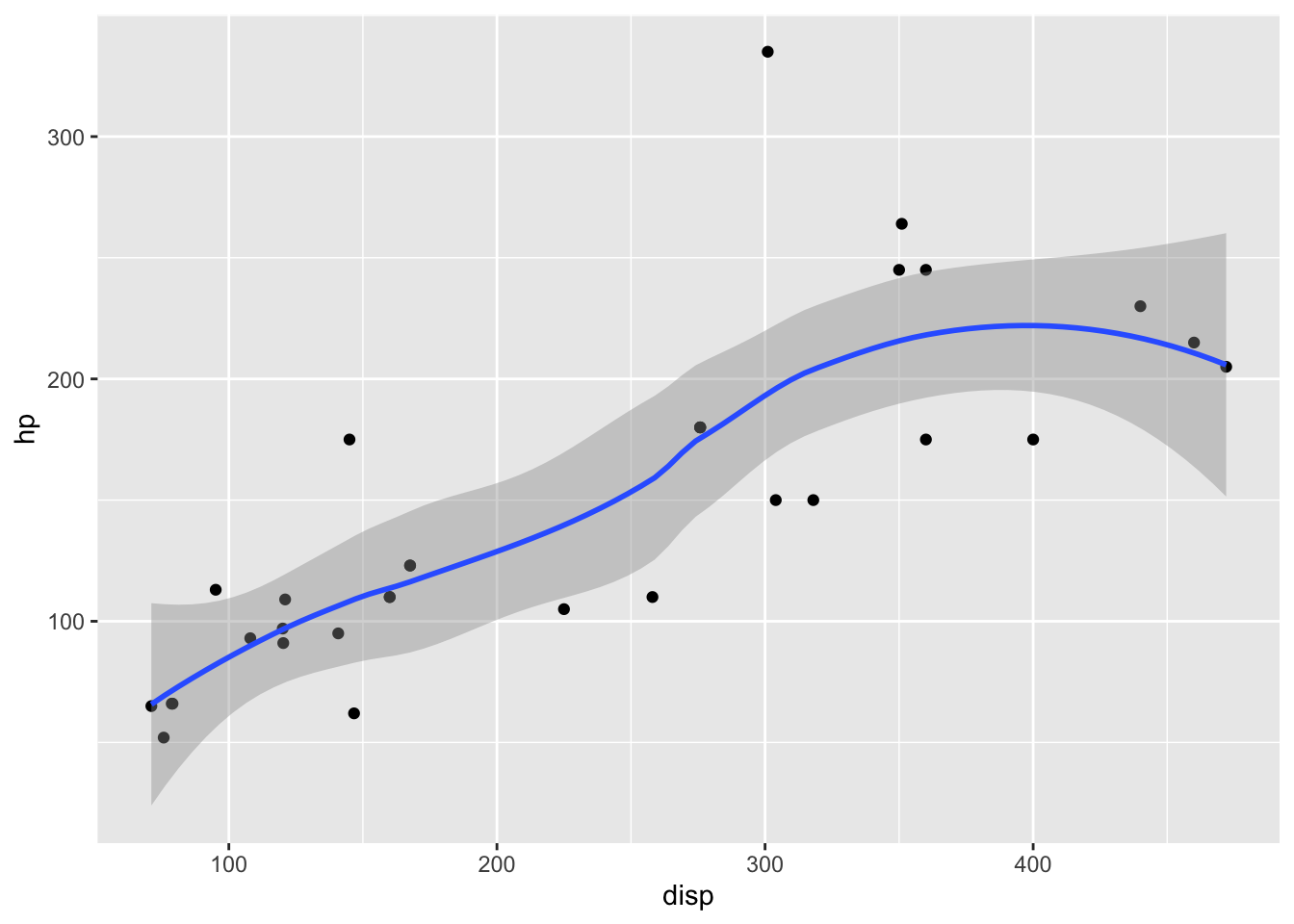
ex <- plot_vars(mtcars, disp, hp)
cat(deparse(ex))## ggplot(mtcars, aes(disp, hp)) + geom_point()eval(plot_vars(mtcars, disp, hp))
ex_smooth <- plot_vars(mtcars, disp, hp, smooth = TRUE)
cat(deparse(ex_smooth))## ggplot(mtcars, aes(disp, hp)) + geom_point() + geom_smooth()eval(ex_smooth)## `geom_smooth()` using method = 'loess' and formula 'y ~ x'